




Dematin rabbit pAb
 One-click to copy product information
One-click to copy product information$148.00/50µL $248.00/100µL
| 50 µL | $148.00 |
| 100 µL | $248.00 |
Overview
| Product name: | Dematin rabbit pAb |
| Reactivity: | Human;Mouse |
| Alternative Names: | EPB49; DMT; Dematin; Erythrocyte membrane protein band 4.9 |
| Source: | Rabbit |
| Dilutions: | Western Blot: 1/500 - 1/2000. Immunohistochemistry: 1/100 - 1/300. Immunofluorescence: 1/200 - 1/1000. ELISA: 1/5000. Not yet tested in other applications. |
| Immunogen: | The antiserum was produced against synthesized peptide derived from human Dematin. AA range:356-405 |
| Storage: | -20°C/1 year |
| Clonality: | Polyclonal |
| Isotype: | IgG |
| Concentration: | 1 mg/ml |
| Observed Band: | 50kD |
| GeneID: | 2039 |
| Human Swiss-Prot No: | Q08495 |
| Cellular localization: | Cytoplasm. Cytoplasm, cytosol. Cytoplasm, perinuclear region . Cytoplasm, cytoskeleton. Cell membrane. Membrane . Endomembrane system. Cell projection . Localized at the spectrin-actin junction of erythrocyte plasma membrane. Localized to intracellular membranes and the cytoskeletal network. Localized at intracellular membrane-bounded organelle compartment in platelets that likely represent the dense tubular network membrane. Detected at the cell membrane and at the parasitophorous vacuole in malaria-infected erythrocytes at late stages of plasmodium berghei or falciparum development. |
| Background: | The protein encoded by this gene is an actin binding and bundling protein that plays a structural role in erythrocytes, by stabilizing and attaching the spectrin/actin cytoskeleton to the erythrocyte membrane in a phosphorylation-dependent manner. This protein contains a core domain in the N-terminus, and a headpiece domain in the C-terminus that binds F-actin. When purified from erythrocytes, this protein exists as a trimer composed of two 48 kDa polypeptides and a 52 kDa polypeptide. The different subunits arise from alternative splicing in the 3' coding region, where the headpiece domain is located. Disruption of this gene has been correlated with the autosomal dominant Marie Unna hereditary hypotrichosis disease, while loss of heterozygosity of this gene is thought to play a role in prostate cancer progression. Alternative splicing results in multiple transcript variants encoding di |
-
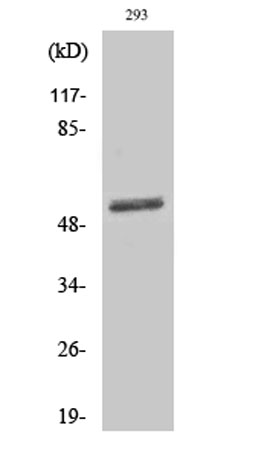
 Western Blot analysis of various cells using Dematin Polyclonal Antibody
Western Blot analysis of various cells using Dematin Polyclonal Antibody -
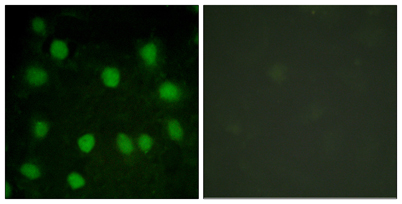
 Immunofluorescence analysis of HUVEC cells, using Dematin Antibody. The picture on the right is blocked with the synthesized peptide.
Immunofluorescence analysis of HUVEC cells, using Dematin Antibody. The picture on the right is blocked with the synthesized peptide. -
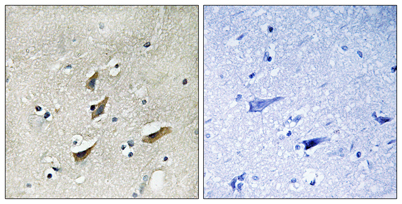
 Immunohistochemistry analysis of paraffin-embedded human brain tissue, using Dematin Antibody. The picture on the right is blocked with the synthesized peptide.
Immunohistochemistry analysis of paraffin-embedded human brain tissue, using Dematin Antibody. The picture on the right is blocked with the synthesized peptide. -
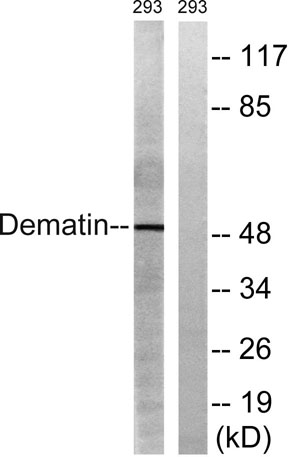
 Western blot analysis of lysates from 293 cells, using Dematin Antibody. The lane on the right is blocked with the synthesized peptide.
Western blot analysis of lysates from 293 cells, using Dematin Antibody. The lane on the right is blocked with the synthesized peptide.

 Manual
Manual