




DDX11 rabbit pAb
 One-click to copy product information
One-click to copy product information$148.00/50µL $248.00/100µL
| 50 µL | $148.00 |
| 100 µL | $248.00 |
Overview
| Product name: | DDX11 rabbit pAb |
| Reactivity: | Human; Mouse |
| Alternative Names: | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.4.13) (CHL1-related protein 1) (hCHLR1) (DEAD/H box protein 11) (Keratinocyte growth factor-regulated gene 2 protein) (KRG-2) |
| Source: | Rabbit |
| Dilutions: | WB 1:500-2000 |
| Immunogen: | Synthesized peptide derived from human DDX11 AA range: 50-100 |
| Storage: | -20°C/1 year |
| Clonality: | Polyclonal |
| Isotype: | IgG |
| Concentration: | 1 mg/ml |
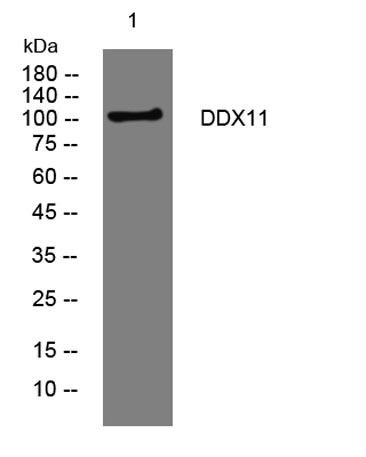
| Observed Band: | 105kD |
| GeneID: | 1663 |
| Human Swiss-Prot No: | Q96FC9 |
| Cellular localization: | Nucleus . Nucleus, nucleolus . Cytoplasm, cytoskeleton, spindle pole . Midbody . Cytoplasm, cytoskeleton, microtubule organizing center, centrosome . During the early stages of mitosis, localizes to condensed chromatin and is released from the chromatin with progression to metaphase. Also localizes to the spindle poles throughout mitosis and at the midbody at later stages of mitosis (metaphase to telophase) (PubMed:17105772). In interphase, colocalizes with nucleolin in the nucleolus (PubMed:26089203). .; Chromosome . (Microbial infection) Colocalizes with bovine papillomavirus type 1 regulatory protein E2 on mitotic chromosomes at early stages of mitosis. . |
| Background: | DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein, which is an enzyme that possesses both ATPase and DNA helicase activities. This gene is a homolog of the yeast CHL1 gene, and may function to maintain chromosome transmission fidelity and genome stability. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Jul 2008], |

 Manual
Manual